ANOVA
One-way anova: aov()
A t-test is used to test hypotheses about the means of two independent samples, for example, to test if there is a difference between control and treatment groups. Two compare more than two independent samples, analysis of variance (ANOVA) is used.
x = c(4, 3, 4, 5, 2, 3, 4, 5)
y = c(4, 4, 5, 5, 4, 5, 4, 4)
z = c(3, 4, 2, 4, 5, 5, 4, 4)
scores = data.frame(x, y, z)
boxplot(scores)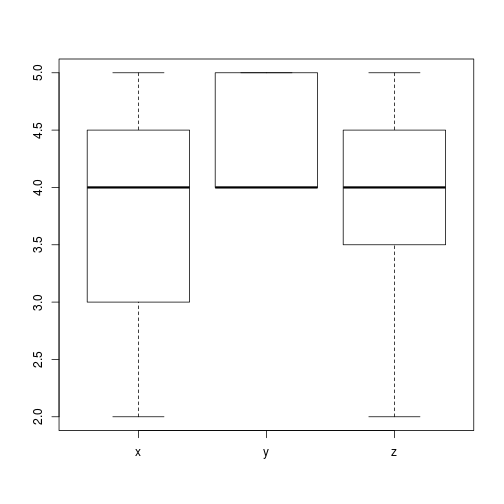
Fig. Boxplot of means
Analysis of variance allows us to investigate if x, y, and z have the same mean. The R function to do the analysis of variance hypothesis test (oneway.test()) requires the data to be in a different format. It wants to have the data with a single variable holding the scores, and a factor describing the grader or category. The stack() command will do this for us:
scores = stack(scores) # look at scores if not clear
names(scores)## [1] "values" "ind"head(scores)## values ind
## 1 4 x
## 2 3 x
## 3 4 x
## 4 5 x
## 5 2 x
## 6 3 xRun the anova using aov()
m <- aov(values ~ ind, data = scores)
summary(m)## Df Sum Sq Mean Sq F value Pr(>F)
## ind 2 1.75 0.875 1.13 0.34
## Residuals 21 16.25 0.774
### Non-parametric anova: kruskal-wallis()kruskal.test(values ~ ind, data = scores)##
## Kruskal-Wallis rank sum test
##
## data: values by ind
## Kruskal-Wallis chi-squared = 1.939, df = 2, p-value = 0.3793Two-way anova
For two independent predictor variables.
gr <- read.table("../data/growth.txt", header = T)Plot the data.
tapply(gr$gain, list(gr$diet, gr$supplement), mean)## agrimore control supergain supersupp
## barley 26.35 23.30 22.47 25.58
## oats 23.30 20.49 19.66 21.86
## wheat 19.64 17.41 17.01 19.67barplot(tapply(gr$gain, list(gr$diet, gr$supplement), mean), beside = TRUE,
ylim = c(0, 30), col = rainbow(3))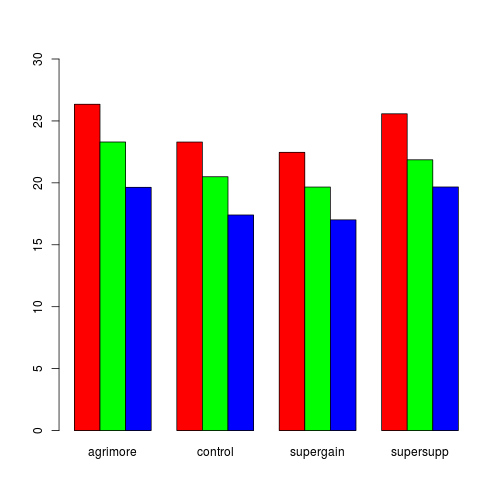
Fig. Growth by diet and supplement
Run the full interaction model
model <- aov(gain ~ diet * supplement, data = gr)
summary(model)## Df Sum Sq Mean Sq F value Pr(>F)
## diet 2 287.2 143.6 83.52 3e-14 ***
## supplement 3 91.9 30.6 17.81 3e-07 ***
## diet:supplement 6 3.4 0.6 0.33 0.92
## Residuals 36 61.9 1.7
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Run the simpler additive model.
model2 <- aov(gain ~ diet + supplement, data = gr)
summary.lm(model2)##
## Call:
## aov(formula = gain ~ diet + supplement, data = gr)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.3079 -0.8593 -0.0771 0.9205 2.9061
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 26.123 0.441 59.26 < 2e-16 ***
## dietoats -3.093 0.441 -7.02 1.4e-08 ***
## dietwheat -5.990 0.441 -13.59 < 2e-16 ***
## supplementcontrol -2.697 0.509 -5.30 4.0e-06 ***
## supplementsupergain -3.381 0.509 -6.64 4.7e-08 ***
## supplementsupersupp -0.727 0.509 -1.43 0.16
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.25 on 42 degrees of freedom
## Multiple R-squared: 0.853, Adjusted R-squared: 0.836
## F-statistic: 48.8 on 5 and 42 DF, p-value: <2e-16We can examine the model in two ways.
- To return an ANOVA table, use
aov()andsummary(). - To return coefficient estimates, use
aov()andsummary.lm(); orlm()andsummary().
We can also use the anova() function to compare the two models, and test for a significant difference between them. Which model is better?
anova(model, model2)## Analysis of Variance Table
##
## Model 1: gain ~ diet * supplement
## Model 2: gain ~ diet + supplement
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 36 61.9
## 2 42 65.3 -6 -3.41 0.33 0.92