Meta-analysis
analysis of analyses
A meta-analysis is the synthesis of
It gives greater weight to studies with less variance and more precision.
NB: A meta-analysis starts with a systematic review.
Effects
An "effect" could be almost any aggregate statistic of interest:
- Mean, Mean difference, Mean change
- Risk ratio, Odds ratio, Risk difference
- Incidence rate, Prevalence, Proportion
- Correlation
Packages
1. Calculate effect sizes
The escalc() function in the metafor package calculates effect sizes.
Provide necessary data components (sample size, events in each treatment group, etc.).
Indicate the effect size you want to measure.
ES <- escalc(endpoints, variances, measure, data, ...)
endpoints: arguments or formula containing endpoint valuesvariances: arguments containing endpoint variancesmeasure: character value indicating type of effect sizedata: data frame containing named variables
The measure (type of effect size) will then influence what endpoint and variance inputs are needed.
See ?escalc for more details.
Example: BCG vaccine trials (from metafor)
Overview: 13 vaccine trials of Bacillus Calmette-Guerin (BCG) vaccine vs. no vaccine
Treatment goal: Prevention of tuberculosis
Primary endpoint: Tuberculosis infection
Possible explanatory variables:
- latitude of study region (
$ablat) - treatment allocation method (
$alloc) - year published (
$year)
library(metafor) # Load package## Loading required package: Matrix
## Loading 'metafor' package (version 1.9-8). For an overview
## and introduction to the package please type: help(metafor).data(dat.bcg) # BCG meta-analytic dataset
str(dat.bcg) # Describe meta-analysis structure## 'data.frame': 13 obs. of 9 variables:
## $ trial : int 1 2 3 4 5 6 7 8 9 10 ...
## $ author: chr "Aronson" "Ferguson & Simes" "Rosenthal et al" "Hart & Sutherland" ...
## $ year : int 1948 1949 1960 1977 1973 1953 1973 1980 1968 1961 ...
## $ tpos : int 4 6 3 62 33 180 8 505 29 17 ...
## $ tneg : int 119 300 228 13536 5036 1361 2537 87886 7470 1699 ...
## $ cpos : int 11 29 11 248 47 372 10 499 45 65 ...
## $ cneg : int 128 274 209 12619 5761 1079 619 87892 7232 1600 ...
## $ ablat : int 44 55 42 52 13 44 19 13 27 42 ...
## $ alloc : chr "random" "random" "random" "random" ...Provides data on the number of TB positive (pos) and negative (neg) patients who were in the treatment (t, given the vaccine) or control (c) group.
Log odds ratios
Compute effect size (yi) and variances (vi) by hand:
Y <- with(dat.bcg, log(tpos * cneg/(tneg * cpos)))
V <- with(dat.bcg, 1/tpos + 1/cneg + 1/tneg + 1/cpos)
cbind(Y, V)## Y V
## [1,] -0.93869414 0.357124952
## [2,] -1.66619073 0.208132394
## [3,] -1.38629436 0.433413078
## [4,] -1.45644355 0.020314413
## [5,] -0.21914109 0.051951777
## [6,] -0.95812204 0.009905266
## [7,] -1.63377584 0.227009675
## [8,] 0.01202060 0.004006962
## [9,] -0.47174604 0.056977124
## [10,] -1.40121014 0.075421726
## [11,] -0.34084965 0.012525134
## [12,] 0.44663468 0.534162172
## [13,] -0.01734187 0.071635117Using escalc, we provide the number of positive and negative patients in the treatment and control groups (i.e., a 2 x 2 frequency table):
ES <- escalc(measure = "OR",
ai = tpos, bi = tneg, ci = cpos, di = cneg,
data = dat.bcg)
cbind(ES$yi, ES$vi)## [,1] [,2]
## [1,] -0.93869414 0.357124952
## [2,] -1.66619073 0.208132394
## [3,] -1.38629436 0.433413078
## [4,] -1.45644355 0.020314413
## [5,] -0.21914109 0.051951777
## [6,] -0.95812204 0.009905266
## [7,] -1.63377584 0.227009675
## [8,] 0.01202060 0.004006962
## [9,] -0.47174604 0.056977124
## [10,] -1.40121014 0.075421726
## [11,] -0.34084965 0.012525134
## [12,] 0.44663468 0.534162172
## [13,] -0.01734187 0.0716351172. Modelling approaches
The function rma() (random effects meta analysis) is a wrapper for escalc and also the main modelling function of metafor. As such, it can also compute effect sizes before modelling.
Fixed
- Is a description of the studies
- Same mean ES, zero between-study variance
Random
- Regards the studies as a sample of a larger universe of studies
- Different mean ES, between-study variance
Random effect model can be used to infer what would likely happen if a new study were performed---Fixed effect model cannot.
Common practice is to report both fixed and random effects model results.
rma()
rma(yi, vi, method, ...)
yieffect sizevivariancesmethodtype of model approach (FE, HE, REML, ...)
fixed effect
Using the hand-calculated ORs
m1 <- rma(yi = Y, vi = V, method = "FE") # Log Odds Ratio
summary(m1)##
## Fixed-Effects Model (k = 13)
##
## logLik deviance AIC BIC AICc
## -76.0290 163.1649 154.0580 154.6229 154.4216
##
## Test for Heterogeneity:
## Q(df = 12) = 163.1649, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## -0.4361 0.0423 -10.3190 <.0001 -0.5190 -0.3533 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Using escalc:
m2 <- rma(method = "FE",
measure = "OR",
ai = tpos, bi = tneg, ci = cpos, di = cneg,
data = dat.bcg)
summary(m2)##
## Fixed-Effects Model (k = 13)
##
## logLik deviance AIC BIC AICc
## -76.0290 163.1649 154.0580 154.6229 154.4216
##
## Test for Heterogeneity:
## Q(df = 12) = 163.1649, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## -0.4361 0.0423 -10.3190 <.0001 -0.5190 -0.3533 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1random effect
m3 <- rma(method = "REML",
measure = "OR",
ai = tpos, bi = tneg, ci = cpos, di = cneg,
data = dat.bcg)
summary(m3)##
## Random-Effects Model (k = 13; tau^2 estimator: REML)
##
## logLik deviance AIC BIC AICc
## -12.5757 25.1513 29.1513 30.1211 30.4847
##
## tau^2 (estimated amount of total heterogeneity): 0.3378 (SE = 0.1784)
## tau (square root of estimated tau^2 value): 0.5812
## I^2 (total heterogeneity / total variability): 92.07%
## H^2 (total variability / sampling variability): 12.61
##
## Test for Heterogeneity:
## Q(df = 12) = 163.1649, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## -0.7452 0.1860 -4.0057 <.0001 -1.1098 -0.3806 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13. The Forest Plot
The forest plot is a plot of effect sizes and their precisions. It is the most common way to report the results of a meta-analysis.
# basic forest plot
forest(m2)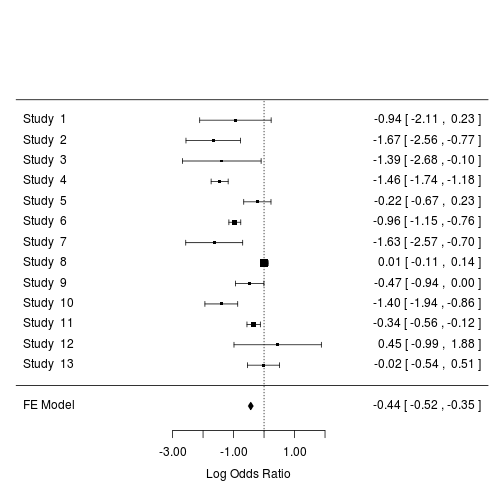
Customising the forest plot:
order: Sort by "obs", "fit", "prec", etc.slab: Change study labelsilab: Add study informationtransf: Apply function to effectspsize: Symbol sizes
Order the studies by precision and transform log odds ratio to odds ratio:
forest(m2, order = "prec", transf = exp, refline = 1)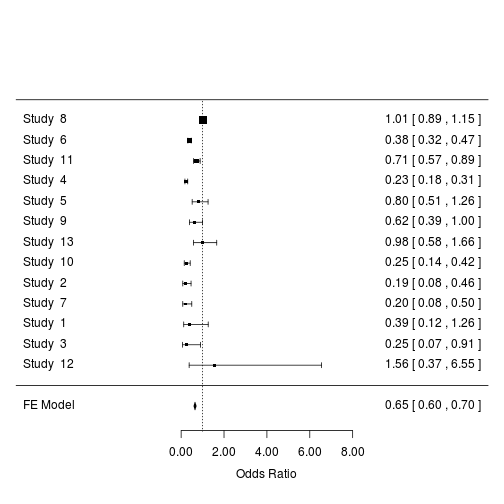
Add the sample size and year of the studies:
# sample size column to data
dat.bcg$n <- with(dat.bcg, tpos + tneg + cpos + cneg)
# plot
forest(m2, order = "prec",
ilab = dat.bcg[, c("n", "year")],
ilab.xpos = exp(m2$b) - c(4, 2),
transf = exp, refline = 1)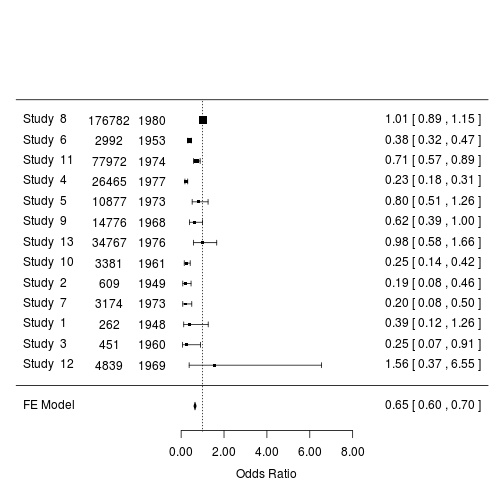
4. Meta-Regression With rma
Meta-regression is used to specify study covariates through the mods argument of rma.
The mods argument takes a matrix of p covariates.
m4 <- rma(ai = tpos, bi = tneg, ci = cpos, di = cneg,
data = dat.bcg,
mods = dat.bcg[, "ablat"],
measure = "OR",
method = "DL")
summary(m4)##
## Mixed-Effects Model (k = 13; tau^2 estimator: DL)
##
## logLik deviance AIC BIC AICc
## -7.4987 26.1043 20.9974 22.6922 23.6640
##
## tau^2 (estimated amount of residual heterogeneity): 0.0480 (SE = 0.0451)
## tau (square root of estimated tau^2 value): 0.2191
## I^2 (residual heterogeneity / unaccounted variability): 56.17%
## H^2 (unaccounted variability / sampling variability): 2.28
## R^2 (amount of heterogeneity accounted for): 86.90%
##
## Test for Residual Heterogeneity:
## QE(df = 11) = 25.0954, p-val = 0.0088
##
## Test of Moderators (coefficient(s) 2):
## QM(df = 1) = 26.1628, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## intrcpt 0.3030 0.2109 1.4370 0.1507 -0.1103 0.7163
## mods -0.0316 0.0062 -5.1150 <.0001 -0.0437 -0.0195 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 15. Publication bias
It is possible that studies showing a significant intervention effect are more often published than studies with null results (the 'file drawer' problem).
When a meta-analysis is based only on studies reported in the literature, null studies relegated to the file-drawer could bias the summary intervention effect in the direction of efficacy
A funnel plot is a scatter plot of the intervention effect estimates against a measure of study precision.
Asymmetry (gaps) in the funnel may be indicative of publication bias.
Some authors argue that judging asymmetry is too subjective to be useful.
Spurious asymmetry can result from heterogeneity or when ESs are correlated with precision
funnel(m3)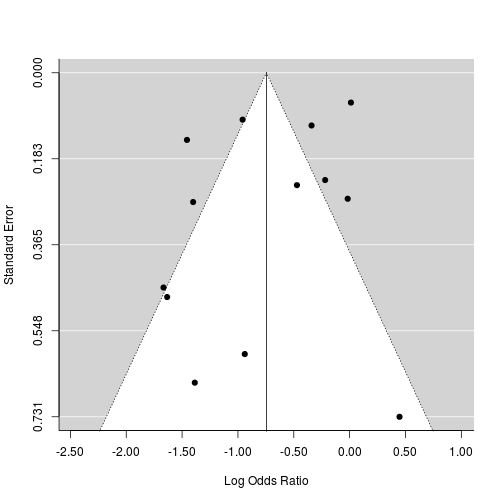
Acknowledgements
http://rpubs.com/skoval/meta
Links
https://ecologyforacrowdedplanet.wordpress.com/2013/05/10/using-metafor-and-ggplot-togetherpart-1/
http://ecologyforacrowdedplanet.wordpress.com/2013/05/30/using-metafor-and-ggplot-togetherpart-2/